Timeline
A selection of our recent work.
-
December 2023
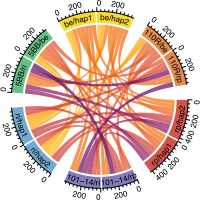
North American wild grapes | Pangenomics
We constructed a super-pangenome to represent and analyze North American wild species of the Vitis genus. The assembly of phased diploid genomes for the nine selected species was a fundamental starting point to ensure an accurate representation of genetic variations that occur within and between genomes. We were able to navigate through different layers of information integrated within the pangenome graph from large structural variations to single-nucleotide polymorphisms. Read the article.
-

November 2023
Grape trunk pathogens | Pangenomics
We sequenced fifty isolates from viticulture regions worldwide and built nucleotide-level, reference-free pangenomes for each species. Through examining genomic diversity and pangenome structure, we analyzed intraspecific conservation and variability of putative virulence factors, focusing on functions under positive selection, and recent gene-family dynamics of contraction and expansion. Read the article.
-
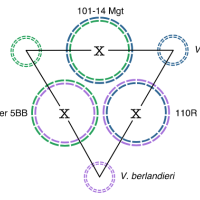
October 2022
Release of three rootstock genomes | HiFi genomics
we report the diploid chromosome-scale assembly of three widely used rootstocks derived from these species: Richter 110 (110R), Kober 5BB, and 101–14 Millardet et de Grasset (Mgt). Read the article.
-
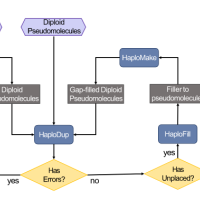
June 2022
Diploid genome scaffolding | Tool release
We developed HaploSync, a suite of tools that produces fully phased, chromosome-scale diploid genome assemblies, and performs extensive quality control to limit assembly artifacts. Read the article.
-

February 2021
Trayshed genome release | Phased genomics with hybrid scaffolds
We report the phased, chromosome-scale assembly of Trayshed, which was produced improving the previous SMRT assembly with the introduction of the optical maps in a hybrid scaffolding approach and consensus genetic map from multiple wild and cultivated grape species. Read the article.
-

June 2020
The genetic basis of sex determination in grapes | Chromosome-scale genomics
We report an improved, chromosome-scale Cabernet Sauvignon genome sequence and the phased assembly of nine wild and cultivated grape genomes. By resolving twenty Vitis SDR haplotypes, we compare male, female, and hermaphrodite haplotype structures and identify sex-linked regions. Read the article.
-
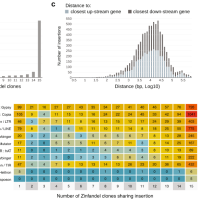
December 2019
Zinfandel clonal diversity
The purpose of this study was to better appreciate clone diversity and involved defining the nature of somatic mutations throughout the genome. Fifteen Zinfandel winegrape clone genomes were sequenced and compared to one another using a highly contiguous genome reference produced from one of the clones, Zinfandel 03. Read the article.
-

Grapegenomics.com release
Release of our community resource website grapegenomics.com with blast and genome browser tools.
-
May 2019
Carménère genome release
Read the article.
-

March 2019
Cabernet Sauvignon Iso-seq | Full-length transcriptomics
We used single molecule-real time sequencing (SMRT) to sequence full-length cDNA (Iso-Seq) and reconstruct the transcriptome of Cabernet Sauvignon berries during berry ripening. Read the article.
-

2019
The Grape Genome | Book release
Read the book.
-

February 2017
Red blotch characterization | Integrated transcriptomics and metabolomics
We integrated genome-wide transcriptional profiling, targeted chemical and biochemical analyses, and demonstrated that grapevine red blotch disrupts ripening and metabolism of red-skinned berries. Read the article.
-

Grapevine trunk pathogens transcriptomics | Metatranscriptomics
Using naturally infected field samples expressing a variety of trunk disease symptoms, we show that our approach provides quantitative assessments of species composition, as well as genomewide transcriptional profiling of potential virulence factors, namely cell wall degradation, secondary metabolism and nutrient uptake for all co-infecting GTPs. Read the article.
-
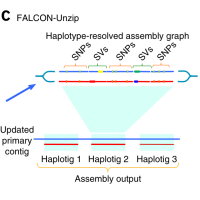
December 2016
Cabernet Sauvignon genome release | Phased genomics
Vitis vinifera cv. Cabernet Sauvignon was selected as a perenial plant model for the release of Falcon and Falcon-Unzip. Read the article.
-

December 2015
Noble rot characterization | Integrated transcriptomics and metabolomics
We integrated transcriptomics, metabolomics, and enzyme activity assays to characterize the impact of noble rot on the development and metabolism of white-skinned grape berries under field conditions. Read the article.
-

June 2015
Grapevine trunk pathogens genomics | Comparative genomics
This study describes the repertoires of putative virulence functions in the genomes of ubiquitous grapevine trunk pathogens. The integration of RNA-seq, comparative and ab initio approaches improved the protein-coding gene prediction in T. minima, whereas shotgun sequencing yielded nearly complete genome drafts of Dia. ampelina, Dip. seriata, and P. chlamydospora. Read the article.
-

December 2014
Powdery mildew genomics | Comparative genomics
A shotgun approach was applied to sequence and assemble the genome of five E. necator isolates, and RNA-seq and comparative genomics were used to predict and annotate protein-coding genes. Read the article.










